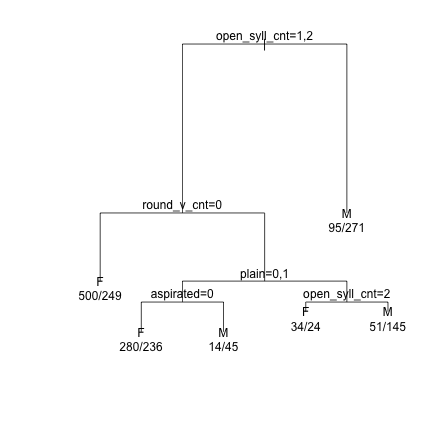
########################################################### ########################################################### ############ PHONETIC ANALYSIS OF KOREAN NAMES ############ ########################################################### ###################### LISA SULLIVAN ###################### ###################### July 13, 2020 ###################### ########################################################### ########################################################### ###################################################### ############ READ FILES FOR ANALYSIS ################# ###################################################### read.delim("phonetic_full_coded_2020_05_07.txt",header=T,as.is = T, comment.char = "#" )->dat_phonetic_full read.delim("phonetic_s1_coded_2020_05_07.txt",header=T,as.is = T, comment.char = "#" )->dat_phonetic_s1 read.delim("phonetic_s2_coded_2020_05_07.txt",header=T,as.is = T, comment.char = "#" )->dat_phonetic_s2 ############################################ ############ CART ANALYSIS ################# ############################################ ######## Factor Everything ########## dat_phonetic_full$open_syll_cnt=factor(dat_phonetic_full$open_syll_cnt) dat_phonetic_full$initial_snd_type=factor(dat_phonetic_full$initial_snd_type) dat_phonetic_full$final_snd_type=factor(dat_phonetic_full$final_snd_type) dat_phonetic_full$syll_1_type=factor(dat_phonetic_full$syll_1_type) dat_phonetic_full$dark_v_cnt=factor(dat_phonetic_full$dark_v_cnt) dat_phonetic_full$light_v_cnt=factor(dat_phonetic_full$light_v_cnt) dat_phonetic_full$front_v_cnt=factor(dat_phonetic_full$front_v_cnt) dat_phonetic_full$back_v_cnt=factor(dat_phonetic_full$back_v_cnt) dat_phonetic_full$high_v_cnt=factor(dat_phonetic_full$high_v_cnt) dat_phonetic_full$low_v_cnt=factor(dat_phonetic_full$low_v_cnt) dat_phonetic_full$round_v_cnt=factor(dat_phonetic_full$round_v_cnt) dat_phonetic_full$diphthongs=factor(dat_phonetic_full$diphthongs) dat_phonetic_full$w_cnt=factor(dat_phonetic_full$w_cnt) dat_phonetic_full$j_cnt=factor(dat_phonetic_full$j_cnt) dat_phonetic_full$stops=factor(dat_phonetic_full$stops) dat_phonetic_full$sonorants=factor(dat_phonetic_full$sonorants) dat_phonetic_full$aspirated=factor(dat_phonetic_full$aspirated) dat_phonetic_full$tense=factor(dat_phonetic_full$tense) dat_phonetic_full$plain=factor(dat_phonetic_full$plain) dat_phonetic_full$Sex=factor(dat_phonetic_full$Sex) dat_phonetic_s1$initial_snd_type=factor(dat_phonetic_s1$initial_snd_type) dat_phonetic_s1$syll_1_type=factor(dat_phonetic_s1$syll_1_type) dat_phonetic_s1$dark_v_cnt=factor(dat_phonetic_s1$dark_v_cnt) dat_phonetic_s1$light_v_cnt=factor(dat_phonetic_s1$light_v_cnt) dat_phonetic_s1$front_v_cnt=factor(dat_phonetic_s1$front_v_cnt) dat_phonetic_s1$back_v_cnt=factor(dat_phonetic_s1$back_v_cnt) dat_phonetic_s1$high_v_cnt=factor(dat_phonetic_s1$high_v_cnt) dat_phonetic_s1$low_v_cnt=factor(dat_phonetic_s1$low_v_cnt) dat_phonetic_s1$round_v_cnt=factor(dat_phonetic_s1$round_v_cnt) dat_phonetic_s1$diphthongs=factor(dat_phonetic_s1$diphthongs) dat_phonetic_s1$w_cnt=factor(dat_phonetic_s1$w_cnt) dat_phonetic_s1$j_cnt=factor(dat_phonetic_s1$j_cnt) dat_phonetic_s1$stops=factor(dat_phonetic_s1$stops) dat_phonetic_s1$sonorants=factor(dat_phonetic_s1$sonorants) dat_phonetic_s1$aspirated=factor(dat_phonetic_s1$aspirated) dat_phonetic_s1$tense=factor(dat_phonetic_s1$tense) dat_phonetic_s1$plain=factor(dat_phonetic_s1$plain) dat_phonetic_s1$Sex=factor(dat_phonetic_s1$Sex) dat_phonetic_s1$height=factor(dat_phonetic_s1$height) dat_phonetic_s1$backness=factor(dat_phonetic_s1$backness) dat_phonetic_s1$dark_bright=factor(dat_phonetic_s1$dark_bright) dat_phonetic_s1$vowel_length=factor(dat_phonetic_s1$vowel_length) dat_phonetic_s2$initial_snd_type=factor(dat_phonetic_s2$initial_snd_type) dat_phonetic_s2$syll_2_type=factor(dat_phonetic_s2$syll_2_type) dat_phonetic_s2$dark_v_cnt=factor(dat_phonetic_s2$dark_v_cnt) dat_phonetic_s2$light_v_cnt=factor(dat_phonetic_s2$light_v_cnt) dat_phonetic_s2$front_v_cnt=factor(dat_phonetic_s2$front_v_cnt) dat_phonetic_s2$back_v_cnt=factor(dat_phonetic_s2$back_v_cnt) dat_phonetic_s2$high_v_cnt=factor(dat_phonetic_s2$high_v_cnt) dat_phonetic_s2$low_v_cnt=factor(dat_phonetic_s2$low_v_cnt) dat_phonetic_s2$round_v_cnt=factor(dat_phonetic_s2$round_v_cnt) dat_phonetic_s2$diphthongs=factor(dat_phonetic_s2$diphthongs) dat_phonetic_s2$w_cnt=factor(dat_phonetic_s2$w_cnt) dat_phonetic_s2$j_cnt=factor(dat_phonetic_s2$j_cnt) dat_phonetic_s2$stops=factor(dat_phonetic_s2$stops) dat_phonetic_s2$sonorants=factor(dat_phonetic_s2$sonorants) dat_phonetic_s2$aspirated=factor(dat_phonetic_s2$aspirated) dat_phonetic_s2$tense=factor(dat_phonetic_s2$tense) dat_phonetic_s2$plain=factor(dat_phonetic_s2$plain) dat_phonetic_s2$Sex=factor(dat_phonetic_s2$Sex) dat_phonetic_s2$height=factor(dat_phonetic_s2$height) dat_phonetic_s2$backness=factor(dat_phonetic_s2$backness) dat_phonetic_s2$dark_bright=factor(dat_phonetic_s2$dark_bright) dat_phonetic_s2$vowel_length=factor(dat_phonetic_s2$vowel_length) ######## Full Names ######## ### Construct & plot the full tree demtree_2s = rpart(Sex~open_syll_cnt+dark_v_cnt+light_v_cnt+front_v_cnt+back_v_cnt+high_v_cnt+low_v_cnt+round_v_cnt+diphthongs+w_cnt+j_cnt+stops+sonorants+aspirated+tense+plain, data = dat_phonetic_full) plot(demtree_2s, compress= T, branch = 1, margin = 0.1) text(demtree_2s, use.n=T,pretty=0)
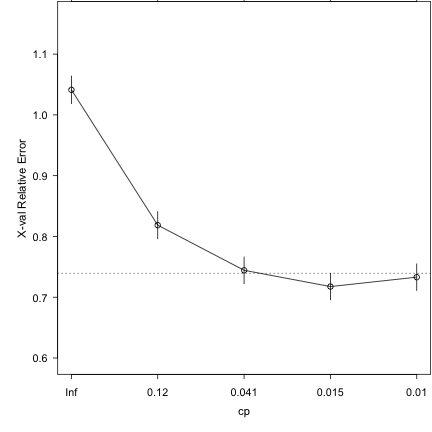
### Run 10-part cross-validation to prune the full tree plotcp(demtree_2s)
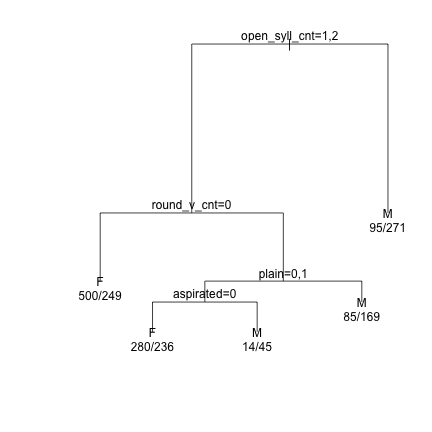
### Prune the tree and replot the data demtree_2s = prune(demtree_2s, cp = 0.015) plot(demtree_2s, compress = T, branch = 1, margin = 0.1) text(demtree_2s, use.n=T,pretty=0)
### Get an estimate of the predictive power of the tree based on the current data predict(demtree_2s, type = "class") -> predValues_2s #Predicted Values preds_2s = data.frame(obs=dat_phonetic_full$Sex, pred=predValues_2s) #Observed vs Predicted xtabs(~obs+pred, preds_2s) #Cross-tabs of observed vs predicted
## pred ## obs F M ## F 780 194 ## M 485 485
(780+485)/nrow(dat_phonetic_full)# Proportion correct
## [1] 0.6507202
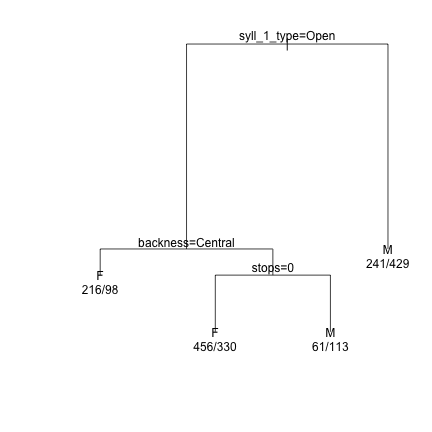
######## Syllable 1 ######## demtree_s1 = rpart(Sex~syll_1_type+initial_snd_type+dark_bright+backness+height+round_v_cnt+vowel_length+stops+sonorants+aspirated+tense+plain, data = dat_phonetic_s1) plot(demtree_s1, compress= T, branch = 1, margin = 0.1) text(demtree_s1, use.n=T,pretty=0)
### Run 10-part cross-validation to prune the full tree plotcp(demtree_s1)
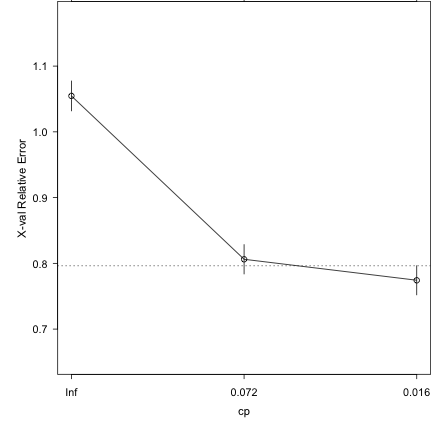
### Prune the tree and replot the data demtree_s1 = prune(demtree_s1, cp = 0.016) plot(demtree_s1, compress = T, branch = 1, margin = 0.1) text(demtree_s1, use.n=T,pretty=0)
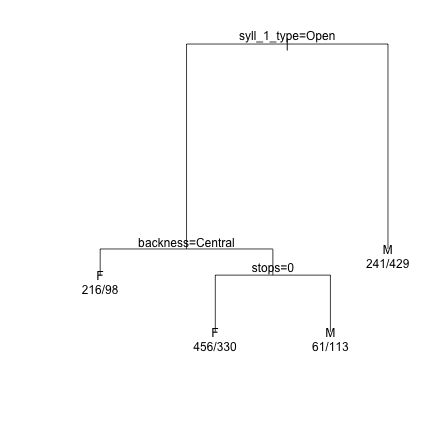
### Get an estimate of the predictive power of the tree based on the current data predict(demtree_s1, type = "class") -> predValues_s1 #Predicted Values preds_s1 = data.frame(obs=dat_phonetic_s1$Sex, pred=predValues_s1) #Observed vs Predicted xtabs(~obs+pred, preds_s1) #Cross-tabs of observed vs predicted
## pred ## obs F M ## F 672 302 ## M 428 542
(672+542)/nrow(dat_phonetic_s1)# Proportion correct
## [1] 0.6244856
######## Syllable 2 ######## demtree_s2 = rpart(Sex~syll_2_type+initial_snd_type+dark_bright+backness+height+round_v_cnt+vowel_length+stops+sonorants+aspirated+tense+plain, data = dat_phonetic_s2) plot(demtree_s2, compress= T, branch = 1, margin = 0.1) text(demtree_s2, use.n=T,pretty=0)
### Run 10-part cross-validation to prune the full tree plotcp(demtree_s2)
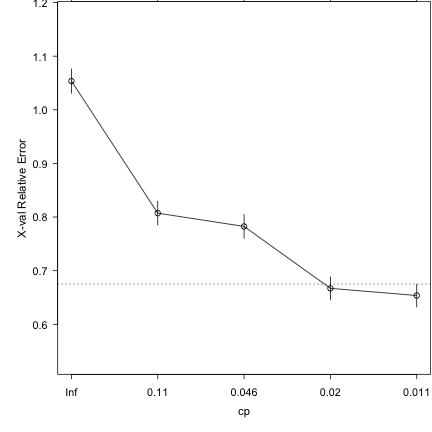
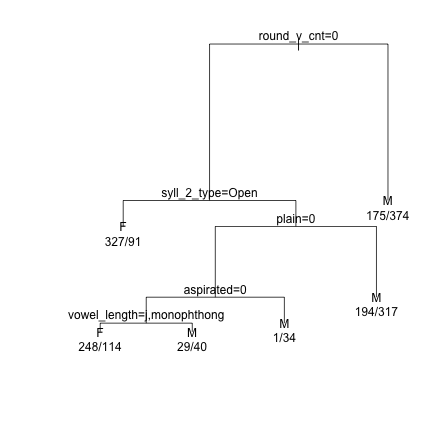
### Prune the tree and replot the data demtree_s2 = prune(demtree_s2, cp = 0.02) plot(demtree_s2, compress = T, branch = 1, margin = 0.1) text(demtree_s2, use.n=T,pretty=0)
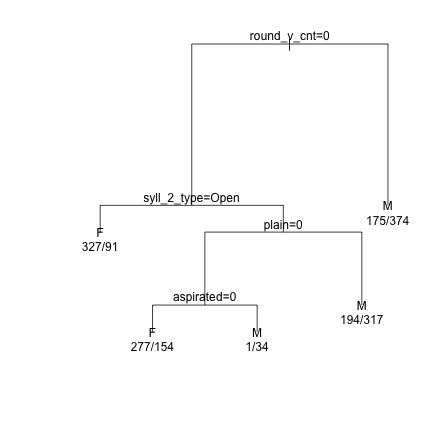
### Get an estimate of the predictive power of the tree based on the current data predict(demtree_s2, type = "class") -> predValues_s2 #Predicted Values preds_s2 = data.frame(obs=dat_phonetic_s2$Sex, pred=predValues_s2) #Observed vs Predicted xtabs(~obs+pred, preds_s2) #Cross-tabs of observed vs predicted
## pred ## obs F M ## F 604 370 ## M 245 725
(604+725)/nrow(dat_phonetic_s2)# Proportion correct
## [1] 0.683642
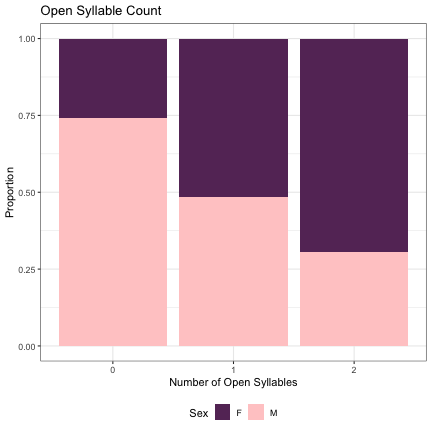
####################################################### ################# UNIVARITE ANALYSIS ################## ####################################################### ############ Code Variables ############# contrasts(dat_phonetic_full$open_syll_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$initial_snd_type)=contrasts(dat_phonetic_full$initial_snd_type)-1/2 contrasts(dat_phonetic_full$final_snd_type)=contrasts(dat_phonetic_full$final_snd_type)-1/2 contrasts(dat_phonetic_full$syll_1_type)=contrasts(dat_phonetic_full$syll_1_type)-1/2 contrasts(dat_phonetic_full$dark_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$light_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$front_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$back_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$high_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$low_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$round_v_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$diphthongs)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$w_cnt)=contrasts(dat_phonetic_full$w_cnt)-1/2 contrasts(dat_phonetic_full$j_cnt)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$stops)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_full$sonorants)=cbind(zero=c(3/4,-1/4,-1/4,-1/4), one=c(0,2/3,-1/3,-1/3), two_three=c(0,0,1/2,-1/2)) contrasts(dat_phonetic_full$aspirated)=contrasts(dat_phonetic_full$aspirated)-1/2 contrasts(dat_phonetic_full$tense)=contrasts(dat_phonetic_full$tense)-1/2 contrasts(dat_phonetic_full$plain)=cbind(zero=c(3/4,-1/4,-1/4,-1/4), one=c(0,2/3,-1/3,-1/3), two_three=c(0,0,1/2,-1/2)) contrasts(dat_phonetic_s1$initial_snd_type)=contrasts(dat_phonetic_s1$initial_snd_type)-1/2 contrasts(dat_phonetic_s1$syll_1_type)=contrasts(dat_phonetic_s1$syll_1_type)-1/2 contrasts(dat_phonetic_s1$dark_v_cnt)=contrasts(dat_phonetic_s1$dark_v_cnt)-1/2 contrasts(dat_phonetic_s1$light_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$front_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$back_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$high_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$low_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$round_v_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$diphthongs)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$w_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$j_cnt)=contrasts(dat_phonetic_s1$light_v_cnt)-1/2 contrasts(dat_phonetic_s1$stops)=contrasts(dat_phonetic_s1$stops)-1/2 contrasts(dat_phonetic_s1$sonorants)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_s1$aspirated)=contrasts(dat_phonetic_s1$aspirated)-1/2 ###contrasts(dat_phonetic_s1$tense)=contrasts(dat_phonetic_s1$tense)-1/2 contrasts(dat_phonetic_s1$plain)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_s1$height)=cbind(high=c(2/3,-1/3,-1/3), low_mid=c(0,1/2,-1/2)) contrasts(dat_phonetic_s1$backness)=cbind(front=c(-1/3,-1/3,2/3), back_central=c(1/2,-1/2,0)) contrasts(dat_phonetic_s1$dark_bright)=cbind(neutral=c(-1/3,-1/3,2/3), bright_dark=c(1/2,-1/2,0)) contrasts(dat_phonetic_s1$vowel_length)=cbind(mono_diph=c(-1/3,2/3,-1/3), j_w=c(1/2,0,-1/2)) ##### ******** NO TENSE IN SYLL 1 contrasts(dat_phonetic_s2$initial_snd_type)=contrasts(dat_phonetic_s2$initial_snd_type)-1/2 contrasts(dat_phonetic_s2$syll_2_type)=contrasts(dat_phonetic_s2$syll_2_type)-1/2 contrasts(dat_phonetic_s2$dark_v_cnt)=contrasts(dat_phonetic_s2$dark_v_cnt)-1/2 contrasts(dat_phonetic_s2$light_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$front_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$back_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$high_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$low_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$round_v_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$diphthongs)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$w_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$j_cnt)=contrasts(dat_phonetic_s2$light_v_cnt)-1/2 contrasts(dat_phonetic_s2$stops)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_s2$sonorants)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_s2$aspirated)=contrasts(dat_phonetic_s2$aspirated)-1/2 contrasts(dat_phonetic_s2$tense)=contrasts(dat_phonetic_s2$tense)-1/2 contrasts(dat_phonetic_s2$plain)=cbind(zero=c(2/3,-1/3,-1/3), one_two=c(0,1/2,-1/2)) contrasts(dat_phonetic_s2$height)=cbind(high=c(2/3,-1/3,-1/3), low_mid=c(0,1/2,-1/2)) contrasts(dat_phonetic_s2$backness)=cbind(front=c(-1/3,-1/3,2/3), back_central=c(1/2,-1/2,0)) contrasts(dat_phonetic_s2$dark_bright)=cbind(neutral=c(-1/3,-1/3,2/3), bright_dark=c(1/2,-1/2,0)) contrasts(dat_phonetic_s2$vowel_length)=cbind(mono_diph=c(-1/3,2/3,-1/3), j_w=c(1/2,0,-1/2)) ######### Full Names ######## # Number of syllables lm_2s_syll_type = glmer(Sex~open_syll_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_syll_type)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ open_syll_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1786.8 1814.7 -888.4 1776.8 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.5981 -0.4009 -0.0309 0.3806 2.6769 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.328 2.516 ## syllable_1 (Intercept) 2.570 1.603 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.4654 0.3113 1.495 0.135 ## open_syll_cntzero 2.2746 0.4987 4.561 5.08e-06 *** ## open_syll_cntone_two 1.6282 0.3839 4.241 2.22e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) opn_s_ ## opn_syll_cn 0.007 ## opn_syll_c_ -0.104 0.771
dat_phonetic_full %>% ggplot(aes(open_syll_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Open Syllables") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Open Syllable Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
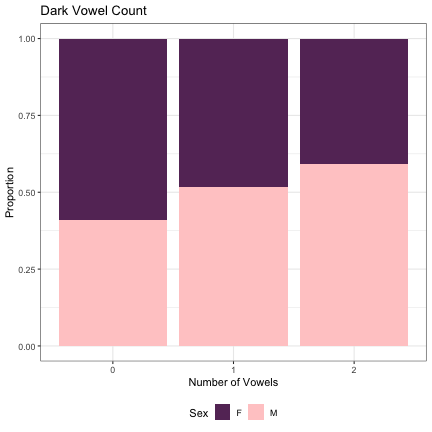
# Number of Dark Vowels lm_2s_dark_v = glmer(Sex~dark_v_cnt + (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_dark_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ dark_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1802.8 1830.7 -896.4 1792.8 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4067 -0.4053 -0.0355 0.3802 2.6514 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.465 2.543 ## syllable_1 (Intercept) 3.119 1.766 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.6843 0.3102 2.206 0.02740 * ## dark_v_cntzero -1.2406 0.4795 -2.587 0.00967 ** ## dark_v_cntone_two -0.7949 0.3588 -2.215 0.02673 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) drk_v_ ## drk_v_cntzr -0.168 ## drk_v_cntn_ -0.192 0.848
dat_phonetic_full %>% ggplot(aes(dark_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Dark Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
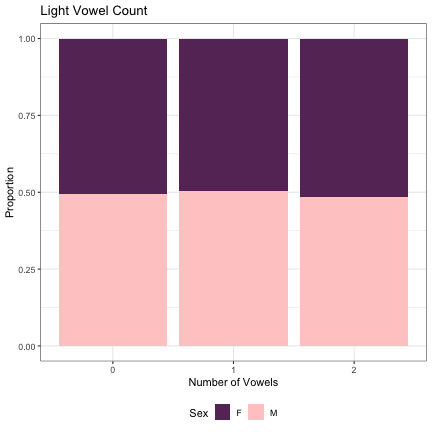
# Number of Light Vowels lm_2s_light_v= glmer(Sex~light_v_cnt + (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_light_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ light_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1809.2 1837.1 -899.6 1799.2 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4394 -0.4088 -0.0375 0.3826 2.5776 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.028 2.651 ## syllable_1 (Intercept) 2.969 1.723 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.55500 0.32342 1.716 0.0862 . ## light_v_cntzero 0.12130 0.47772 0.254 0.7996 ## light_v_cntone_two 0.03229 0.43573 0.074 0.9409 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) lght__ ## lght_v_cntz -0.267 ## lght_v_cnt_ -0.303 0.803
dat_phonetic_full %>% ggplot(aes(light_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Light Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
# Number of Front Vowels lm_2s_front_v = glmer(Sex~front_v_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_front_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ front_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1798.0 1825.9 -894.0 1788.0 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.5658 -0.4095 -0.0313 0.3807 2.4007 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.073 2.464 ## syllable_1 (Intercept) 3.205 1.790 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.07682 0.35493 -0.216 0.828652 ## front_v_cntzero 1.71998 0.55373 3.106 0.001895 ** ## front_v_cntone_two 1.51670 0.45001 3.370 0.000751 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) frnt__ ## frnt_v_cntz -0.512 ## frnt_v_cnt_ -0.509 0.859
dat_phonetic_full %>% ggplot(aes(front_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Front Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
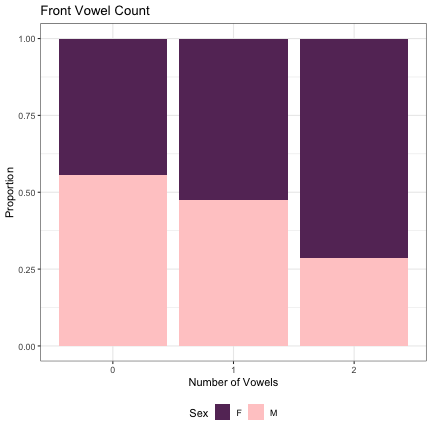
# Number of Back Vowels lm_2s_back_v = glmer(Sex~back_v_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_back_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ back_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1800.5 1828.4 -895.3 1790.5 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.3124 -0.4038 -0.0355 0.3822 2.6481 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.248 2.500 ## syllable_1 (Intercept) 3.074 1.753 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.8836 0.3459 2.555 0.01062 * ## back_v_cntzero -1.4284 0.5321 -2.684 0.00727 ** ## back_v_cntone_two -0.7206 0.5036 -1.431 0.15246 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) bck_v_ ## bck_v_cntzr -0.457 ## bck_v_cntn_ -0.481 0.829
dat_phonetic_full %>% ggplot(aes(back_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Back Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
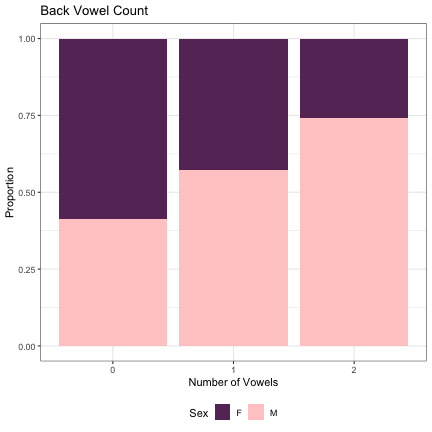
# Number of High Vowels lm_2s_high_v = glmer(Sex~high_v_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_high_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ high_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1809.3 1837.2 -899.7 1799.3 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4244 -0.4090 -0.0373 0.3826 2.5648 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.025 2.65 ## syllable_1 (Intercept) 2.958 1.72 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.57710 0.32020 1.802 0.0715 . ## high_v_cntzero -0.05076 0.48666 -0.104 0.9169 ## high_v_cntone_two -0.04318 0.35552 -0.121 0.9033 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) hgh_v_ ## hgh_v_cntzr -0.243 ## hgh_v_cntn_ -0.276 0.845
dat_phonetic_full %>% ggplot(aes(high_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("High Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
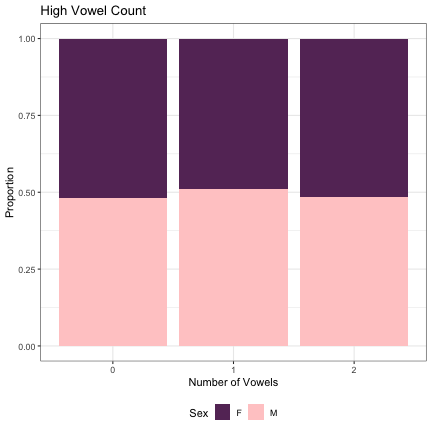
# Number of Low Vowels lm_2s_low_v = glmer(Sex~low_v_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_low_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ low_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1808.9 1836.8 -899.5 1798.9 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4217 -0.4071 -0.0369 0.3807 2.5961 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.968 2.640 ## syllable_1 (Intercept) 3.000 1.732 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.4285 0.3813 1.124 0.261 ## low_v_cntzero 0.3718 0.5668 0.656 0.512 ## low_v_cntone_two 0.2641 0.5961 0.443 0.658 ## ## Correlation of Fixed Effects: ## (Intr) lw_v_c ## low_v_cntzr -0.573 ## lw_v_cntn_t -0.543 0.799
dat_phonetic_full %>% ggplot(aes(low_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Low Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
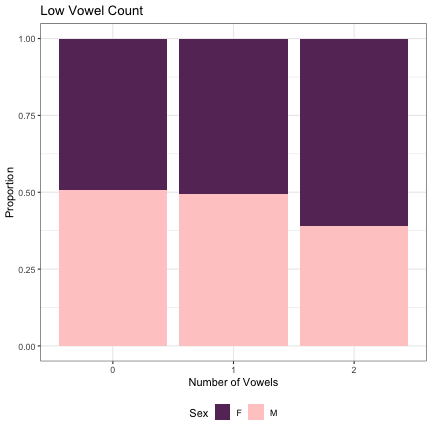
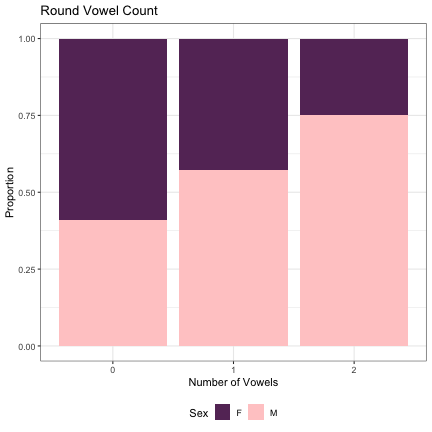
# Number of Round Vowels lm_2s_round_v = glmer(Sex~round_v_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_round_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ round_v_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1800.3 1828.2 -895.1 1790.3 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.2930 -0.4033 -0.0356 0.3829 2.6477 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.228 2.496 ## syllable_1 (Intercept) 3.073 1.753 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.8840 0.3435 2.573 0.01007 * ## round_v_cntzero -1.4569 0.5288 -2.755 0.00587 ** ## round_v_cntone_two -0.7596 0.4993 -1.521 0.12821 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) rnd_v_ ## rnd_v_cntzr -0.445 ## rnd_v_cntn_ -0.473 0.828
dat_phonetic_full %>% ggplot(aes(round_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Round Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
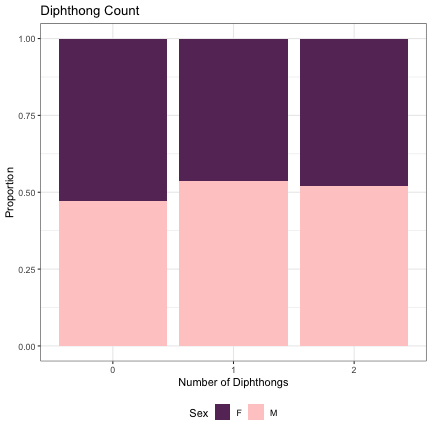
# Number of Diphthongs lm_2s_diphthong = glmer(Sex~diphthongs+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_diphthong)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ diphthongs + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1807.9 1835.8 -898.9 1797.9 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.3921 -0.4068 -0.0368 0.3815 2.5729 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.884 2.624 ## syllable_1 (Intercept) 2.997 1.731 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.7545 0.3808 1.981 0.0475 * ## diphthongszero -0.5969 0.5688 -1.050 0.2939 ## diphthongsone_two -0.2748 0.5493 -0.500 0.6169 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) dphthn ## diphthngszr -0.572 ## dphthngsn_t -0.557 0.821
dat_phonetic_full %>% ggplot(aes(diphthongs,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Diphthongs") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Diphthong Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
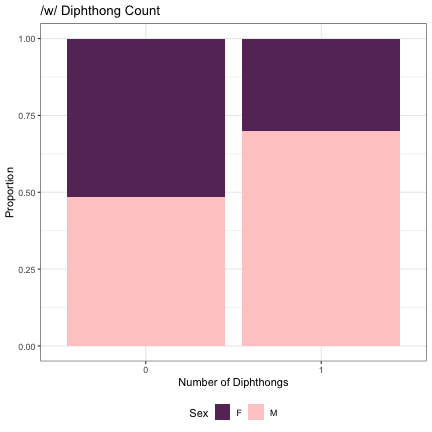
# Number of /w/ Diphthongs lm_2s_w = glmer(Sex~w_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_w)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ w_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1800.7 1822.9 -896.3 1792.7 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.3901 -0.4111 -0.0375 0.3812 2.5532 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.939 2.634 ## syllable_1 (Intercept) 2.862 1.692 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.4478 0.4789 3.023 0.0025 ** ## w_cnt1 2.0876 0.8282 2.521 0.0117 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## w_cnt1 0.744
dat_phonetic_full %>% ggplot(aes(w_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Diphthongs") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("/w/ Diphthong Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
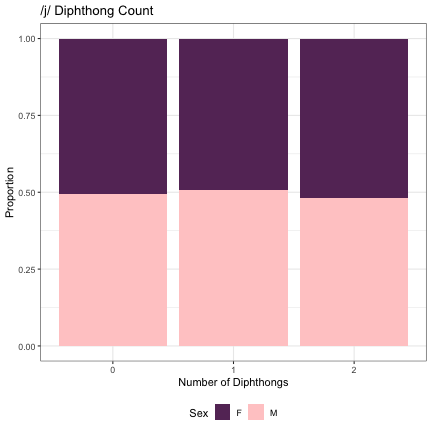
# Number of /j/ Diphthongs lm_2s_j = glmer(Sex~j_cnt+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_j)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ j_cnt + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1809.3 1837.2 -899.7 1799.3 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4316 -0.4106 -0.0374 0.3841 2.5609 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.029 2.651 ## syllable_1 (Intercept) 2.951 1.718 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.57286 0.42537 1.347 0.178 ## j_cntzero 0.01982 0.63544 0.031 0.975 ## j_cntone_two -0.06890 0.66447 -0.104 0.917 ## ## Correlation of Fixed Effects: ## (Intr) j_cntz ## j_cntzero -0.673 ## j_cntone_tw -0.646 0.833
dat_phonetic_full %>% ggplot(aes(j_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Diphthongs") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("/j/ Diphthong Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
# Number of Stops lm_2s_stops = glmer(Sex~stops+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_stops)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ stops + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1787.5 1815.4 -888.8 1777.5 1939 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.2202 -0.4104 -0.0357 0.3759 2.6083 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.504 2.550 ## syllable_1 (Intercept) 2.747 1.657 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.3370 0.3842 3.480 0.000502 *** ## stopszero -2.3427 0.5666 -4.135 3.55e-05 *** ## stopsone_two -1.1910 0.5786 -2.058 0.039548 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) stpszr ## stopszero -0.571 ## stopsone_tw -0.526 0.787
dat_phonetic_full %>% ggplot(aes(stops,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Stops") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Stop Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
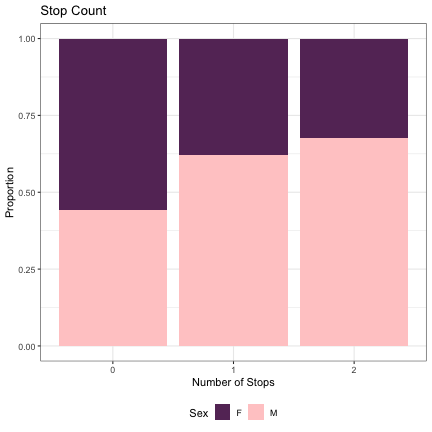
# Number of Sonorants lm_2s_sonorants = glmer(Sex~sonorants+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_sonorants)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ sonorants + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1807.5 1840.9 -897.7 1795.5 1938 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.5447 -0.4025 -0.0379 0.3781 2.6041 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.079 2.661 ## syllable_1 (Intercept) 2.826 1.681 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.5557 0.3281 1.694 0.0903 . ## sonorantszero -0.9948 0.5699 -1.746 0.0809 . ## sonorantsone -0.4024 0.4546 -0.885 0.3761 ## sonorantstwo_three -0.4019 0.5062 -0.794 0.4273 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) snrntsz snrntsn ## sonorantszr -0.126 ## sonorantson -0.273 0.828 ## snrntstw_th -0.322 0.600 0.779
dat_phonetic_full %>% ggplot(aes(sonorants,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Sonorants") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Sonorant Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
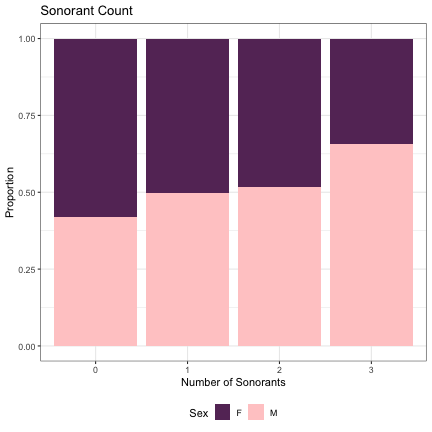
# Number of Lenis Obstruents lm_2s_plain = glmer(Sex~plain+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_plain)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ plain + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1801.6 1835.1 -894.8 1789.6 1938 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.2792 -0.4058 -0.0368 0.3783 2.7033 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.262 2.502 ## syllable_1 (Intercept) 2.911 1.706 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.8833 0.4060 2.175 0.02960 * ## plainzero -1.9483 0.6561 -2.969 0.00298 ** ## plainone -1.4007 0.6879 -2.036 0.04174 * ## plaintwo_three -1.1669 1.1821 -0.987 0.32357 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) planzr plainn ## plainzero -0.463 ## plainone -0.634 0.854 ## plaintw_thr -0.670 0.636 0.883
dat_phonetic_full %>% ggplot(aes(plain,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Lenis Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
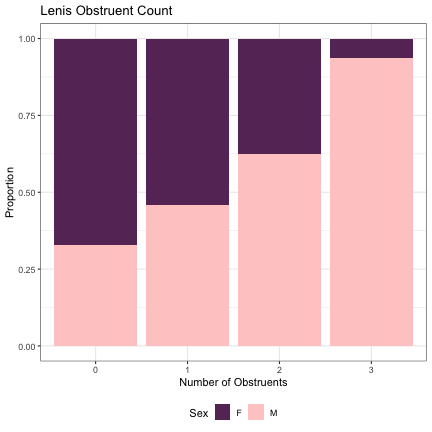
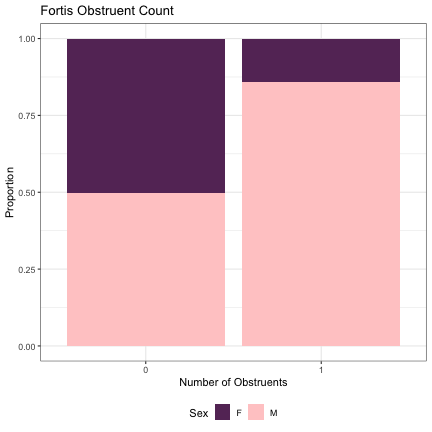
# Number of Fortis Obstruents lm_2s_tense = glmer(Sex~tense+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_tense)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ tense + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1806.4 1828.7 -899.2 1798.4 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4275 -0.4087 -0.0372 0.3840 2.5576 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.126 2.669 ## syllable_1 (Intercept) 2.903 1.704 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.608 1.064 1.512 0.131 ## tense1 2.152 2.113 1.018 0.308 ## ## Correlation of Fixed Effects: ## (Intr) ## tense1 0.953
dat_phonetic_full %>% ggplot(aes(tense,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Fortis Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
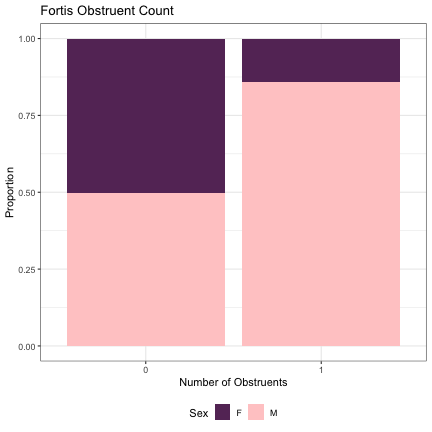
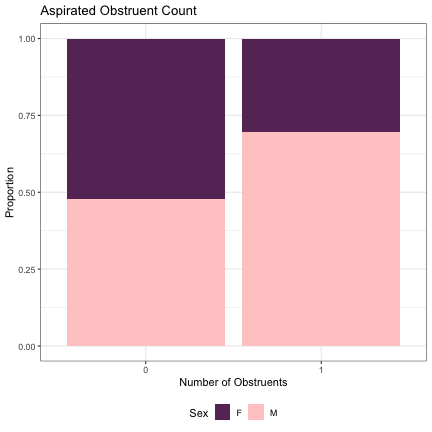
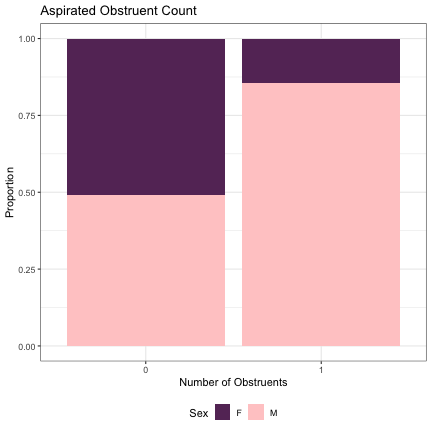
# Number of Aspirated Obstruents lm_2s_aspirated = glmer(Sex~aspirated+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_aspirated)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ aspirated + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1801.5 1823.8 -896.8 1793.5 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.3971 -0.4079 -0.0357 0.3807 2.5699 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.927 2.632 ## syllable_1 (Intercept) 2.924 1.710 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.2480 0.4319 2.890 0.00386 ** ## aspirated1 1.6966 0.7135 2.378 0.01742 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## aspirated1 0.671
dat_phonetic_full %>% ggplot(aes(aspirated,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Aspirated Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
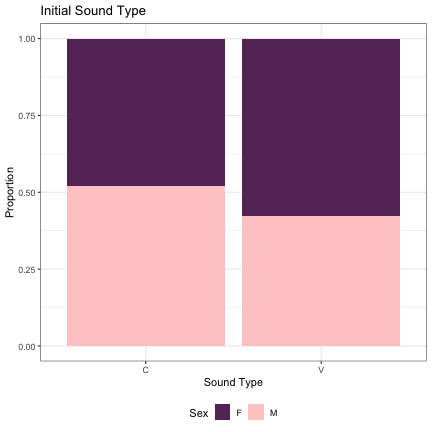
######### Syllable #1 ########## # Initial Sound lm_s1_initial_snd = glmer(Sex~initial_snd_type + (1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_initial_snd)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ initial_snd_type + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2390.2 2407.0 -1192.1 2384.2 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9526 -0.7651 -0.2140 0.7768 3.0933 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.456 1.567 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.0946 0.2187 -0.433 0.665 ## initial_snd_typeV -0.2564 0.4370 -0.587 0.557 ## ## Correlation of Fixed Effects: ## (Intr) ## intl_snd_tV 0.613
dat_phonetic_s1 %>% ggplot(aes(initial_snd_type,fill=Sex)) + geom_bar(position="fill") + xlab("Sound Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Initial Sound Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
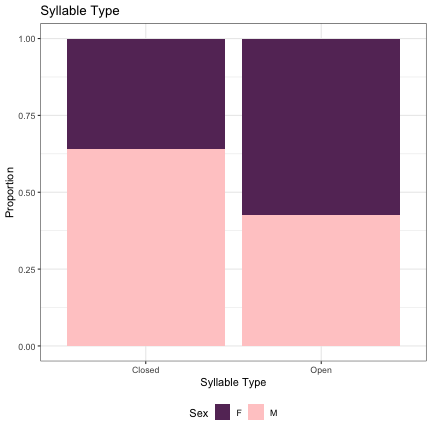
# Syllable Type lm_s1_initial_syll = glmer(Sex~syll_1_type+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_initial_syll)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ syll_1_type + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2378.6 2395.3 -1186.3 2372.6 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.0074 -0.7626 -0.2165 0.7680 3.1122 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 1.987 1.41 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.1033 0.1619 0.638 0.523605 ## syll_1_typeOpen -1.1673 0.3242 -3.601 0.000317 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## syll_1_typO -0.189
dat_phonetic_s1 %>% ggplot(aes(syll_1_type,fill=Sex)) + geom_bar(position="fill") + xlab("Syllable Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Syllable Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
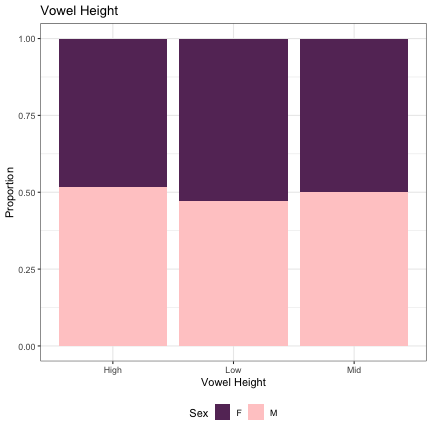
# Vowel Height lm_s1_height = glmer(Sex~height+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_height)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ height + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2391.1 2413.4 -1191.5 2383.1 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9728 -0.7642 -0.2152 0.7755 3.0836 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.462 1.569 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.04983 0.17628 -0.283 0.777 ## heighthigh 0.27831 0.37349 0.745 0.456 ## heightlow_mid -0.45848 0.43275 -1.059 0.289 ## ## Correlation of Fixed Effects: ## (Intr) hghthg ## heighthigh -0.006 ## heightlw_md 0.193 -0.137
dat_phonetic_s1 %>% ggplot(aes(height,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Height") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Height") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
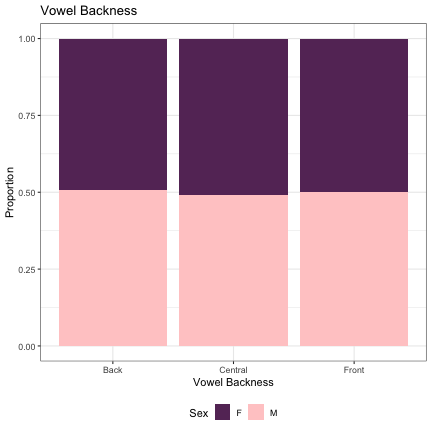
# Vowel Backness lm_s1_backness = glmer(Sex~backness+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_backness)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ backness + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2392.3 2414.6 -1192.2 2384.3 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9360 -0.7633 -0.2116 0.7790 3.1327 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.472 1.572 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.03521 0.17854 -0.197 0.844 ## backnessfront -0.15927 0.39156 -0.407 0.684 ## backnessback_central -0.10287 0.42086 -0.244 0.807 ## ## Correlation of Fixed Effects: ## (Intr) bcknss ## backnssfrnt 0.099 ## bcknssbck_c 0.206 -0.140
dat_phonetic_s1 %>% ggplot(aes(backness,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Backness") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Backness") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
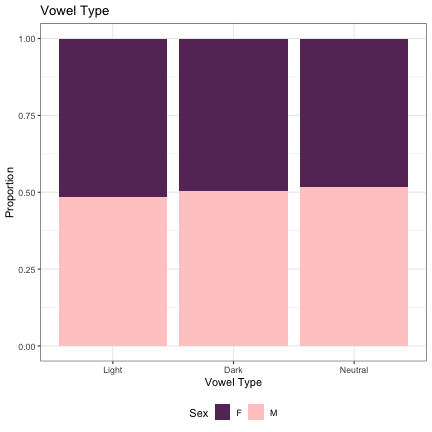
#Vowel Type (Dark - Light - Neutral) lm_s1_darkness = glmer(Sex~dark_bright+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_darkness)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ dark_bright + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2390.1 2412.4 -1191.1 2382.1 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.8357 -0.7670 -0.2046 0.7662 3.0405 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.451 1.565 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.01060 0.18247 -0.058 0.954 ## dark_brightneutral 0.01197 0.43335 0.028 0.978 ## dark_brightbright_dark -0.60969 0.38771 -1.573 0.116 ## ## Correlation of Fixed Effects: ## (Intr) drk_br ## drk_brghtnt 0.323 ## drk_brghtb_ -0.009 0.010
dat_phonetic_s1 -> dat_phonetic_s1b dat_phonetic_s1b$dark_bright = as.character(dat_phonetic_s1b$dark_bright) dat_phonetic_s1b %>% mutate(dark_bright=if_else(dark_bright == "Bright", "Light", dark_bright)) -> dat_phonetic_s1b dat_phonetic_s1b$dark_bright = factor(dat_phonetic_s1b$dark_bright, levels = c("Light", "Dark", "Neutral")) dat_phonetic_s1b %>% ggplot(aes(dark_bright,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
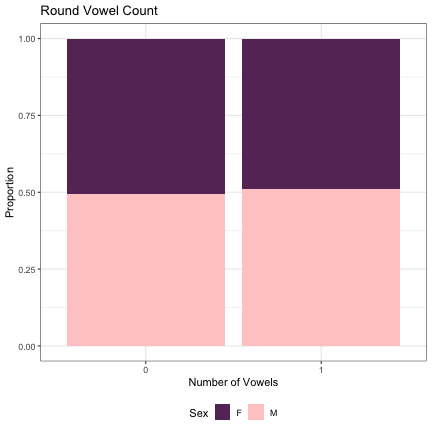
# Vowel Rounding lm_s1_round_v = glmer(Sex~round_v_cnt+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_round_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ round_v_cnt + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2390.5 2407.3 -1192.3 2384.5 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9620 -0.7648 -0.2135 0.7776 3.1117 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.475 1.573 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.03996 0.33068 0.121 0.904 ## round_v_cnt1 0.07763 0.38828 0.200 0.842 ## ## Correlation of Fixed Effects: ## (Intr) ## rond_v_cnt1 0.852
dat_phonetic_s1 %>% ggplot(aes(round_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Round Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
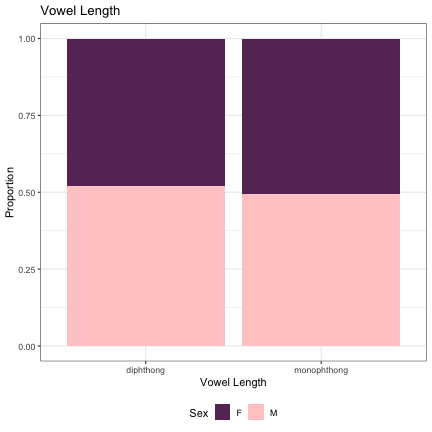
# Vowel Length & Diphthong Type lm_s1_diphthong = glmer(Sex~vowel_length+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_diphthong)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ vowel_length + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2388.6 2410.8 -1190.3 2380.6 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.8704 -0.7702 -0.2126 0.7329 3.1095 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.374 1.541 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.5475 0.3324 1.647 0.0995 . ## vowel_lengthmono_diph -0.9997 0.5264 -1.899 0.0575 . ## vowel_lengthj_w -1.6102 0.9791 -1.645 0.1000 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) vwl_lngthm_ ## vwl_lngthm_ -0.842 ## vwl_lngthj_ -0.650 0.617
dat_phonetic_s1 %>% mutate(vowel_length = if_else(vowel_length == "monophthong", "monophthong", "diphthong")) %>% ggplot(aes(vowel_length,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Length") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Length") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
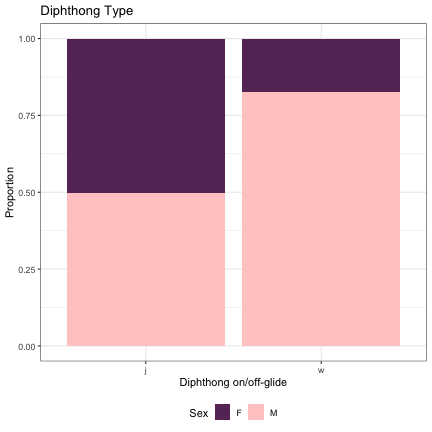
dat_phonetic_s1 %>% filter(vowel_length != "monophthong") %>% ggplot(aes(vowel_length,fill=Sex)) + geom_bar(position="fill") + xlab("Diphthong on/off-glide") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Diphthong Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
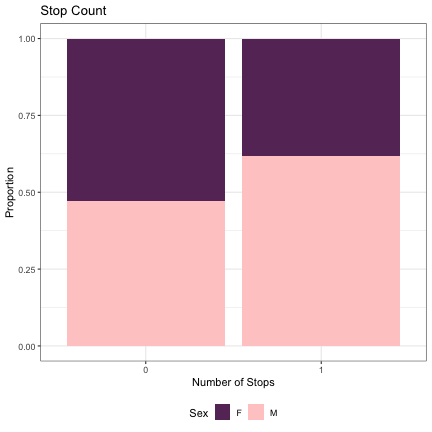
# Number of Stops lm_s1_stops = glmer(Sex~stops+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_stops)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ stops + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2386.4 2403.1 -1190.2 2380.4 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.1115 -0.7686 -0.2129 0.7804 3.1130 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.297 1.516 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.2149 0.2006 1.071 0.2841 ## stops1 0.8383 0.4011 2.090 0.0366 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## stops1 0.543
dat_phonetic_s1 %>% ggplot(aes(stops,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Stops") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Stop Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
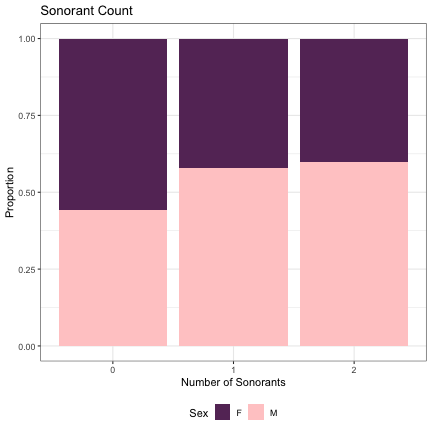
# Number of Sonorants lm_s1_sonorants = glmer(Sex~sonorants+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_sonorants)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ sonorants + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2388.9 2411.2 -1190.5 2380.9 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.0073 -0.7657 -0.2085 0.7797 3.1460 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.323 1.524 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.05232 0.37393 -0.140 0.889 ## sonorantszero -0.41582 0.59639 -0.697 0.486 ## sonorantsone_two 0.49266 1.09524 0.450 0.653 ## ## Correlation of Fixed Effects: ## (Intr) snrnts ## sonorantszr -0.811 ## sonrntsn_tw -0.879 0.824
dat_phonetic_s1 %>% ggplot(aes(sonorants,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Sonorants") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Sonorant Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
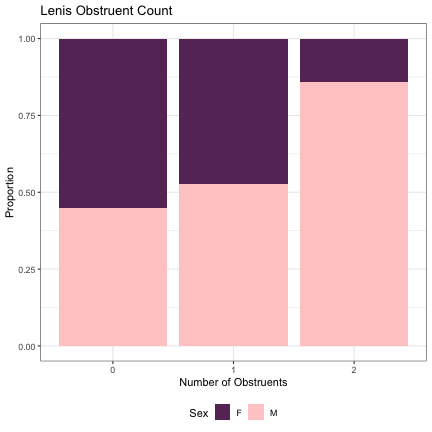
# Number of Lenis Obstruents lm_s1_plain = glmer(Sex~plain+(1|syllable_1) , data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_plain)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ plain + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2390.0 2412.3 -1191.0 2382.0 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9399 -0.7727 -0.2176 0.7804 3.0703 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.384 1.544 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.6338 0.5393 1.175 0.240 ## plainzero -1.2900 0.8433 -1.530 0.126 ## plainone_two -1.9749 1.5954 -1.238 0.216 ## ## Correlation of Fixed Effects: ## (Intr) planzr ## plainzero -0.876 ## plainone_tw -0.947 0.908
dat_phonetic_s1 %>% ggplot(aes(plain,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Lenis Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
# No Fortis Obstruents in S1 # Number of Aspirated Obstruents lm_s1_aspirated = glmer(Sex~aspirated + (1|syllable_1), data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s1_aspirated)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ aspirated + (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2388.6 2405.3 -1191.3 2382.6 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9072 -0.7708 -0.2116 0.7783 3.1130 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 2.436 1.561 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.4132 0.3494 1.183 0.237 ## aspirated1 0.9860 0.7001 1.408 0.159 ## ## Correlation of Fixed Effects: ## (Intr) ## aspirated1 0.870
dat_phonetic_s1 %>% ggplot(aes(aspirated,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Aspirated Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
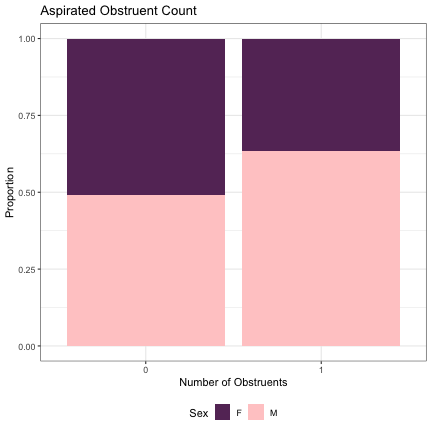
######### Syllable #2 ########## # Initial Sound lm_s2_initial_snd = glmer(Sex~initial_snd_type + (1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_initial_snd)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ initial_snd_type + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2017.3 2034.0 -1005.6 2011.3 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.7487 -0.6135 -0.1371 0.5670 3.3289 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.771 2.602 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.07186 0.29034 0.247 0.805 ## initial_snd_typeV -0.65361 0.58096 -1.125 0.261 ## ## Correlation of Fixed Effects: ## (Intr) ## intl_snd_tV 0.570
dat_phonetic_s2 %>% ggplot(aes(initial_snd_type,fill=Sex)) + geom_bar(position="fill") + xlab("Sound Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Initial Sound Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
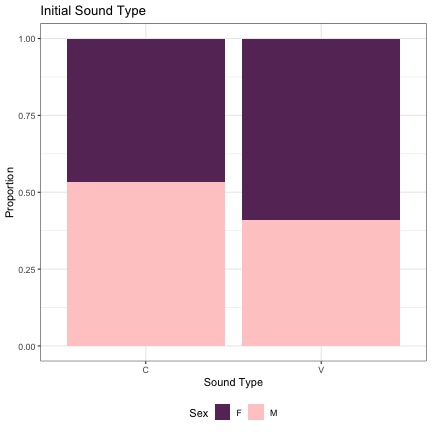
# Syllable Type lm_s2_initial_syll = glmer(Sex~syll_2_type+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_initial_syll)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ syll_2_type + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2010.1 2026.8 -1002.1 2004.1 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.8114 -0.6073 -0.1243 0.5790 3.3708 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.509 2.551 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.02518 0.24714 0.102 0.91885 ## syll_2_typeOpen -1.46275 0.49875 -2.933 0.00336 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## syll_2_typO 0.310
dat_phonetic_s2 %>% ggplot(aes(syll_2_type,fill=Sex)) + geom_bar(position="fill") + xlab("Syllable Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Syllable Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
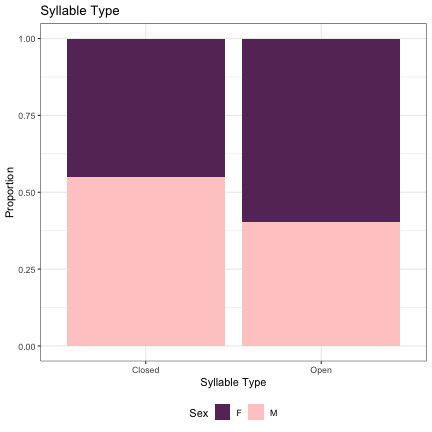
# Vowel Height lm_s2_height = glmer(Sex~height+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_height)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ height + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2018.9 2041.2 -1005.4 2010.9 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.5865 -0.6134 -0.1264 0.5669 3.3436 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.856 2.618 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.1519 0.2528 0.601 0.548 ## heighthigh 0.3722 0.4991 0.746 0.456 ## heightlow_mid -0.7858 0.6620 -1.187 0.235 ## ## Correlation of Fixed Effects: ## (Intr) hghthg ## heighthigh -0.205 ## heightlw_md 0.277 -0.219
dat_phonetic_s2 %>% ggplot(aes(height,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Height") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Height") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
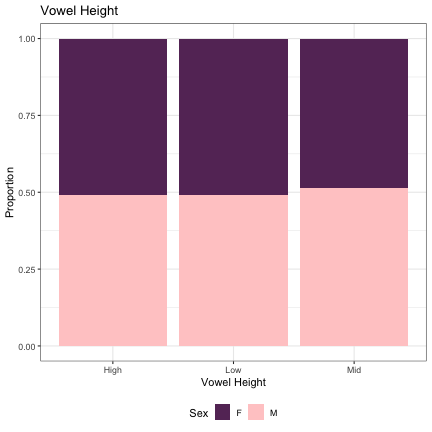
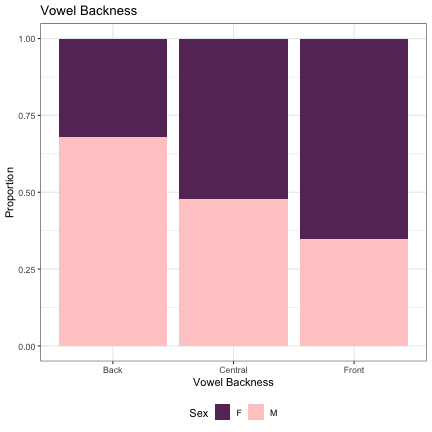
# Vowel Backness lm_s2_backness = glmer(Sex~backness+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_backness)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ backness + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2005.2 2027.4 -998.6 1997.2 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.6220 -0.6224 -0.1431 0.5655 3.5863 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 5.901 2.429 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.1035 0.2414 0.429 0.668189 ## backnessfront -1.9827 0.5620 -3.528 0.000419 *** ## backnessback_central 1.0190 0.5409 1.884 0.059572 . ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) bcknss ## backnssfrnt 0.194 ## bcknssbck_c 0.101 -0.077
dat_phonetic_s2 %>% ggplot(aes(backness,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Backness") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Backness") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
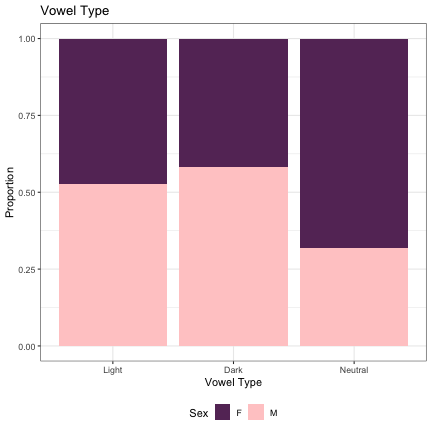
#Vowel Type (Dark - Light - Neutral) lm_s2_darkness = glmer(Sex~dark_bright+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_darkness)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ dark_bright + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2008.8 2031.1 -1000.4 2000.8 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.5663 -0.6202 -0.1384 0.5604 3.5455 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 5.989 2.447 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.001852 0.237678 0.008 0.99378 ## dark_brightneutral -1.529510 0.555312 -2.754 0.00588 ** ## dark_brightbright_dark -1.059477 0.518312 -2.044 0.04094 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) drk_br ## drk_brghtnt 0.274 ## drk_brghtb_ 0.085 -0.055
dat_phonetic_s2 -> dat_phonetic_s2b dat_phonetic_s2b$dark_bright = as.character(dat_phonetic_s2b$dark_bright) dat_phonetic_s2b %>% mutate(dark_bright=if_else(dark_bright == "Bright", "Light", dark_bright)) -> dat_phonetic_s2b dat_phonetic_s2b$dark_bright = factor(dat_phonetic_s2b$dark_bright, levels = c("Light", "Dark", "Neutral")) dat_phonetic_s2b %>% ggplot(aes(dark_bright,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Type") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
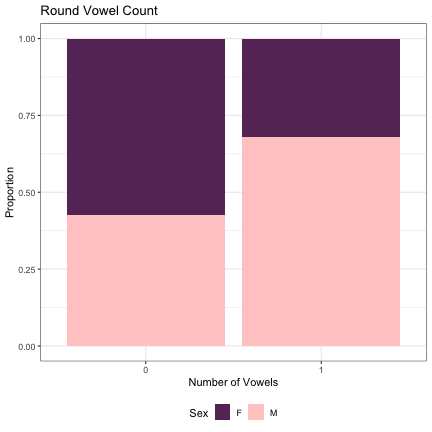
# Vowel Rounding lm_s2_round_v = glmer(Sex~round_v_cnt+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_round_v)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ round_v_cnt + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2008.6 2025.4 -1001.3 2002.6 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.5154 -0.6218 -0.1335 0.5596 3.3994 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.091 2.468 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.2756 0.3928 3.248 0.00116 ** ## round_v_cnt1 1.5669 0.4826 3.247 0.00117 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## rond_v_cnt1 0.813
dat_phonetic_s2 %>% ggplot(aes(round_v_cnt,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Vowels") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Round Vowel Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
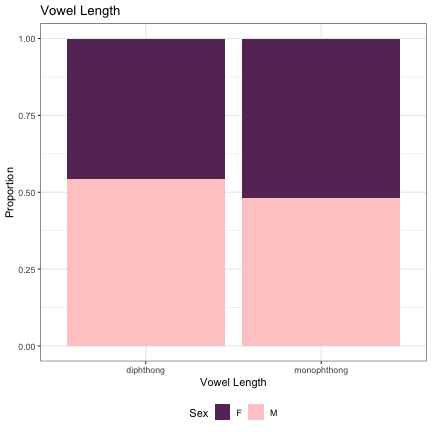
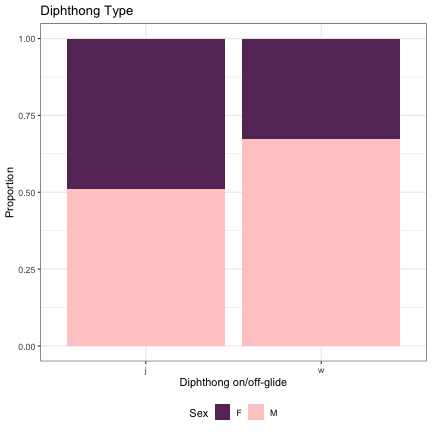
# Vowel Length & Diphthong Type lm_s2_diphthong = glmer(Sex~vowel_length+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_diphthong)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ vowel_length + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2014.0 2036.3 -1003.0 2006.0 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.6455 -0.6104 -0.1322 0.5715 3.3893 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.701 2.589 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.1575 0.4490 2.578 0.00993 ** ## vowel_lengthmono_diph -1.7663 0.7151 -2.470 0.01351 * ## vowel_lengthj_w -2.0526 1.2915 -1.589 0.11199 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) vwl_lngthm_ ## vwl_lngthm_ -0.812 ## vwl_lngthj_ -0.627 0.588
dat_phonetic_s2 %>% mutate(vowel_length = if_else(vowel_length == "monophthong", "monophthong", "diphthong")) %>% ggplot(aes(vowel_length,fill=Sex)) + geom_bar(position="fill") + xlab("Vowel Length") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Vowel Length") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
dat_phonetic_s2 %>% filter(vowel_length != "monophthong") %>% ggplot(aes(vowel_length,fill=Sex)) + geom_bar(position="fill") + xlab("Diphthong on/off-glide") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Diphthong Type") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
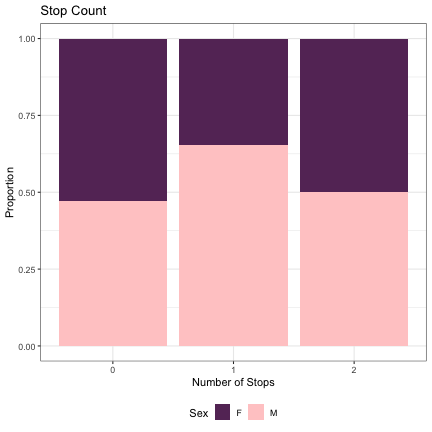
# Number of Stops lm_s2_stops = glmer(Sex~stops+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_stops)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ stops + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2000.5 2022.8 -996.2 1992.5 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.5549 -0.6080 -0.1287 0.5754 3.4438 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.559 2.561 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.9546 0.8273 1.154 0.249 ## stopszero -1.9740 1.2712 -1.553 0.120 ## stopsone_two 1.2541 2.4434 0.513 0.608 ## ## Correlation of Fixed Effects: ## (Intr) stpszr ## stopszero -0.943 ## stopsone_tw -0.897 0.875
dat_phonetic_s2 %>% ggplot(aes(stops,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Stops") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Stop Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
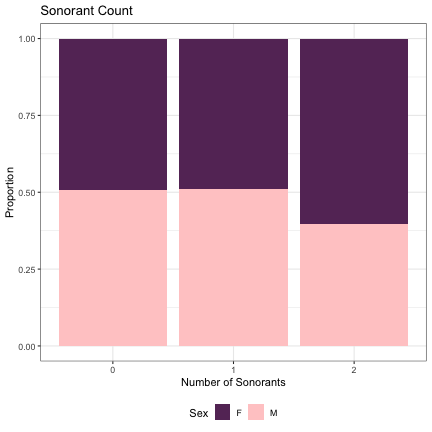
# Number of Sonorants lm_s2_sonorants = glmer(Sex~sonorants+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_sonorants)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ sonorants + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2014.0 2036.3 -1003.0 2006.0 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.7944 -0.6123 -0.1392 0.5647 3.6354 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.688 2.586 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.1405 0.2819 -0.498 0.6183 ## sonorantszero 0.5540 0.5622 0.985 0.3244 ## sonorantsone_two 1.8932 0.7374 2.567 0.0103 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) snrnts ## sonorantszr -0.177 ## sonrntsn_tw -0.536 0.408
dat_phonetic_s2 %>% ggplot(aes(sonorants,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Sonorants") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Sonorant Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
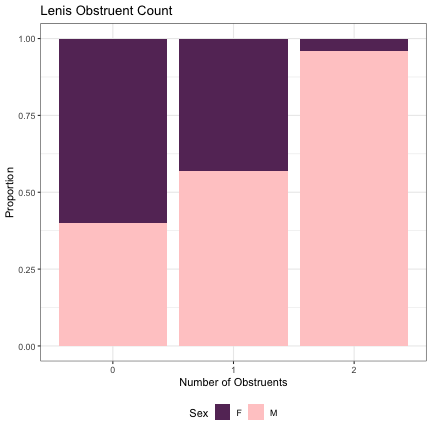
# Number of Lenis Obstruents lm_s2_plain = glmer(Sex~plain+(1|syllable_2) , data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_plain)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ plain + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2004.4 2026.7 -998.2 1996.4 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.4977 -0.6161 -0.1310 0.5672 3.4306 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.178 2.486 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.6144 0.5156 3.131 0.001742 ** ## plainzero -3.0492 0.8297 -3.675 0.000238 *** ## plainone_two -4.0477 1.4962 -2.705 0.006823 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) planzr ## plainzero -0.798 ## plainone_tw -0.872 0.814
dat_phonetic_s2 %>% ggplot(aes(plain,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Lenis Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
# Number of Fortis Obstruents lm_2s_tense = glmer(Sex~tense+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_2s_tense)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ tense + (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1806.4 1828.7 -899.2 1798.4 1940 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.4275 -0.4087 -0.0372 0.3840 2.5576 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 7.126 2.669 ## syllable_1 (Intercept) 2.903 1.704 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.608 1.064 1.512 0.131 ## tense1 2.152 2.113 1.018 0.308 ## ## Correlation of Fixed Effects: ## (Intr) ## tense1 0.953
dat_phonetic_full %>% ggplot(aes(tense,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Fortis Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
# Number of Aspirated Obstruents lm_s2_aspirated = glmer(Sex~aspirated + (1|syllable_2), data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(lm_s2_aspirated)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ aspirated + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2015.2 2031.9 -1004.6 2009.2 1941 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -5.3318 -0.6117 -0.1338 0.5693 3.3629 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 6.819 2.611 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.3464 0.6492 2.074 0.0381 * ## aspirated1 2.3468 1.2920 1.816 0.0693 . ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) ## aspirated1 0.923
dat_phonetic_s2 %>% ggplot(aes(aspirated,fill=Sex)) + geom_bar(position="fill") + xlab("Number of Obstruents") + ylab("Proportion") + theme_bw() + guides(fill=guide_legend(title="Sex")) + ggtitle("Aspirated Obstruent Count") + scale_fill_manual(values=c("#663366","#FFCCCC")) + theme(legend.position="bottom")
########################################## ######### MULTIVARIATE ANALYSIS ########## ########################################## ########### Full name glm_2s_trimmed = glmer(Sex~open_syll_cnt+stops+round_v_cnt+w_cnt+aspirated+ (1|syllable_1) + (1|syllable_2), data=dat_phonetic_full, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(glm_2s_trimmed)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ open_syll_cnt + stops + round_v_cnt + w_cnt + aspirated + ## (1 | syllable_1) + (1 | syllable_2) ## Data: dat_phonetic_full ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1745.2 1806.5 -861.6 1723.2 1933 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.12394 -0.39385 -0.02739 0.37066 2.89442 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 4.619 2.149 ## syllable_1 (Intercept) 2.254 1.501 ## Number of obs: 1944, groups: syllable_2, 163; syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 3.3817 0.6168 5.482 4.20e-08 *** ## open_syll_cntzero 2.3735 0.4747 5.000 5.73e-07 *** ## open_syll_cntone_two 1.6549 0.3670 4.509 6.52e-06 *** ## stopszero -2.1418 0.5255 -4.076 4.59e-05 *** ## stopsone_two -1.0479 0.5593 -1.874 0.06099 . ## round_v_cntzero -2.0938 0.5209 -4.020 5.82e-05 *** ## round_v_cntone_two -1.1612 0.5119 -2.268 0.02331 * ## w_cnt1 2.3789 0.7810 3.046 0.00232 ** ## aspirated1 1.8403 0.6450 2.853 0.00433 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) opn_s_ opn___ stpszr stpsn_ rnd_v_ rnd___ w_cnt1 ## opn_syll_cn 0.069 ## opn_syll_c_ -0.005 0.751 ## stopszero -0.412 0.012 0.014 ## stopsone_tw -0.353 0.001 0.016 0.776 ## rnd_v_cntzr -0.364 -0.176 -0.161 0.084 0.056 ## rnd_v_cntn_ -0.335 -0.111 -0.109 0.053 0.039 0.818 ## w_cnt1 0.628 0.002 -0.009 -0.096 -0.061 -0.091 -0.050 ## aspirated1 0.483 0.069 0.060 -0.012 0.009 -0.105 -0.067 0.065
########### 1st Syllable glm_s1_trimmed = glmer(Sex~syll_1_type+stops+aspirated+vowel_length+height + (1|syllable_1), data=dat_phonetic_s1, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(glm_s1_trimmed)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ syll_1_type + stops + aspirated + vowel_length + height + ## (1 | syllable_1) ## Data: dat_phonetic_s1 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 2375.1 2425.3 -1178.6 2357.1 1935 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.1189 -0.7544 -0.2202 0.7678 3.0884 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_1 (Intercept) 1.62 1.273 ## Number of obs: 1944, groups: syllable_1, 123 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 1.4481 0.4383 3.304 0.000954 *** ## syll_1_typeOpen -1.1376 0.3081 -3.693 0.000222 *** ## stops1 0.8874 0.3600 2.465 0.013701 * ## aspirated1 1.3896 0.6390 2.175 0.029648 * ## vowel_lengthmono_diph -0.8638 0.4872 -1.773 0.076222 . ## vowel_lengthj_w -1.9670 0.9052 -2.173 0.029785 * ## heighthigh 0.5458 0.3272 1.668 0.095257 . ## heightlow_mid -0.5743 0.4112 -1.397 0.162525 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) sy_1_O stops1 asprt1 vwl_lngthm_ vwl_lngthj_ hghthg ## syll_1_typO -0.052 ## stops1 0.246 0.002 ## aspirated1 0.671 -0.035 0.006 ## vwl_lngthm_ -0.660 -0.094 -0.021 -0.092 ## vwl_lngthj_ -0.495 0.026 -0.046 -0.052 0.616 ## heighthigh 0.197 -0.029 0.131 0.178 -0.104 -0.067 ## heightlw_md -0.079 -0.071 -0.064 -0.260 -0.082 0.167 -0.170
########## 2nd Syllable glm_s2_trimmed = glmer(Sex~stops+backness+vowel_length+plain+syll_2_type+aspirated + (1|syllable_2), data=dat_phonetic_s2, family="binomial", control = glmerControl(optimizer = "bobyqa", optCtrl=list(maxfun=2e6))) summary(glm_s2_trimmed)
## Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) [glmerMod ## ] ## Family: binomial ( logit ) ## Formula: Sex ~ stops + backness + vowel_length + plain + syll_2_type + ## aspirated + (1 | syllable_2) ## Data: dat_phonetic_s2 ## Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+06)) ## ## AIC BIC logLik deviance df.resid ## 1967.4 2034.2 -971.7 1943.4 1932 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -4.8161 -0.6232 -0.1210 0.5828 3.6271 ## ## Random effects: ## Groups Name Variance Std.Dev. ## syllable_2 (Intercept) 3.96 1.99 ## Number of obs: 1944, groups: syllable_2, 163 ## ## Fixed effects: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.5165 0.9779 2.573 0.01007 * ## stopszero 7.7263 13.2529 0.583 0.55990 ## stopsone_two 18.1481 26.5103 0.685 0.49362 ## backnessfront -1.4482 0.5261 -2.753 0.00591 ** ## backnessback_central 2.0185 0.5069 3.982 6.83e-05 *** ## vowel_lengthmono_diph -1.5956 0.6148 -2.595 0.00945 ** ## vowel_lengthj_w -3.1816 1.1127 -2.859 0.00425 ** ## plainzero -9.5075 13.2346 -0.718 0.47252 ## plainone_two -17.3870 26.4673 -0.657 0.51123 ## syll_2_typeOpen -1.1714 0.4733 -2.475 0.01332 * ## aspirated1 2.7568 1.1840 2.328 0.01989 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Correlation of Fixed Effects: ## (Intr) stpszr stpsn_ bcknss bckns_ vwl_lngthm_ vwl_lngthj_ planzr plnn_t ## stopszero -0.045 ## stopsone_tw -0.041 0.999 ## backnssfrnt -0.048 0.008 0.005 ## bcknssbck_c 0.081 0.002 0.006 0.042 ## vwl_lngthm_ -0.390 0.002 0.001 -0.144 -0.218 ## vwl_lngthj_ -0.269 0.004 -0.004 0.086 -0.193 0.578 ## plainzero -0.014 -0.997 -0.996 0.002 -0.002 0.002 -0.001 ## plainone_tw -0.012 -0.996 -0.997 0.005 0.001 0.001 0.002 0.999 ## syll_2_typO 0.090 -0.006 -0.003 -0.250 -0.303 0.045 0.094 -0.001 -0.006 ## aspirated1 0.584 -0.004 -0.010 -0.022 0.063 -0.076 -0.016 0.000 0.008 ## sy_2_O ## stopszero ## stopsone_tw ## backnssfrnt ## bcknssbck_c ## vwl_lngthm_ ## vwl_lngthj_ ## plainzero ## plainone_tw ## syll_2_typO ## aspirated1 -0.050